Overview
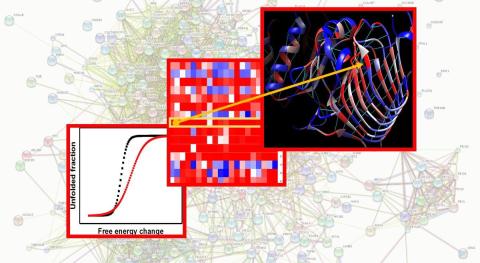
NEI has made the Ocular Proteome dataset available so that researchers can analyze protein changes related to genetic eye diseases. The dataset holds structures for over 100 proteins and over 1 million variants, cataloging the proteins atomic structures and stability generated by global computational mutagenesis.
Resources
Notes
The following people have made important contributions to this project:
- Caitlyn McCafferty wrote a Python code for unfolding mutation screen
- Francisca Wood Ortiz developed a Python code for the protein network
- Mikhail Laryukhin, Divyang Mago, and Samuel Hailai created the Proteome GUI interface
- Yuri Sergeev supervised this project and performed molecular modeling
- Claudia Kassouf, Milan Patel, and Taariq Woods supported database expansion
Questions?
Please reach out to Yuri Sergeev at yuri@nei.gov if you have questions about ocular proteome.